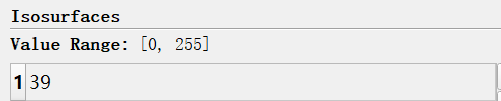
i have a .mha file,then i use vtk.vtkImageMarchingCubes() to get marchingCubes.SetValue(0, 39),the result is bad
but i use Paraview Contour
the result is better than vtk.vtkImageMarchingCubes()。
why?
i have a .mha file,then i use vtk.vtkImageMarchingCubes() to get marchingCubes.SetValue(0, 39),the result is bad
but i use Paraview Contour


the result is better than vtk.vtkImageMarchingCubes()。
why?
ParaView uses vtk.vtkContourFilter which dispatches to a number of different contour filters that perform the actual contouring. To my knowledge, it does not dispatch to vtk.vtkImageMarchingCubes.
Try using vtk.vtkContourFilter in VTK and see how the results match. You should be able to simply replace vtkImageMarchingCubes.
the code is
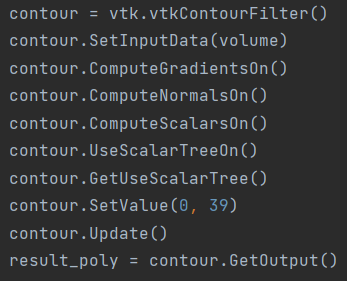
the result is also bad ,just same with vtk.vtkImageMarchingCubes,
Do u have some ideas?
vtk.vtkPVContourFilter is in the ParaView source code, so it isn’t available with just VTK. You could use ParaView’s pvpython application and do your processing there if all you care about is the output isosurface, but I suspect you care about more.
As to why the same isovalue produces different results with the different filters, I’m at a bit of a loss. Is it possible to share your data?
VascularEnhance.rar (808.9 KB)
This is the MRA data。So far, I only care about outputting results because my supervisor asked me to do research on vascular analysis. I use mha(dicom is also ok) to output vascular vtp files, and then use vmtk to generate vascular centerline vtp files。So,I want vascular vtp files to look good
sorry,I know the reason。I downsampled the data ![]()