Voronoi3D tessellation is very expensive for high-resolution data. What I have are dumps from my simulation where cell quantities are defined and the cells are made from 3D Voronoi tessellation. Along with this, I have the data of the face vertex coordinates of the cells and their respective normals and indices. Is it possible to create a generate a Voronoi3D tessellation from this data without the expensive computational overhead in Paraview for visualization?
HI
DO YOU HAVE ANY LEADS ON VISUALIZING THE VORONOI CELL IN PARAVIEW?
Any help will be appreciated!
THANKS
No! But SPH representation gives me an approximate visualization which turned out to be good enough for the Arepo code I was working on.
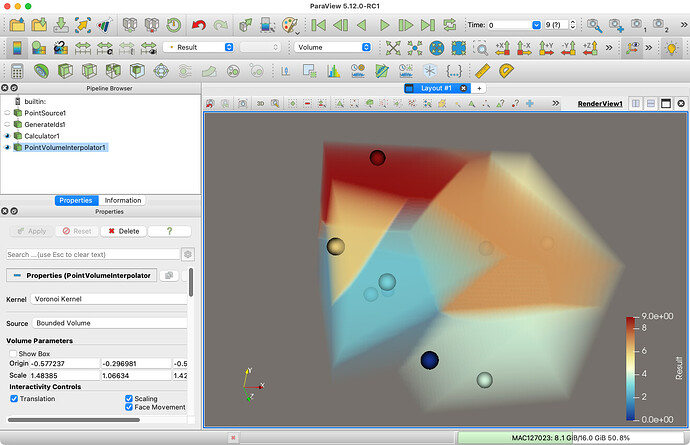
I don’t think there is a way to explicitly create the Voronoi cell tessellation in ParaView. But depending on what you are intending to do with it, you might be able to get away with a sampled Voronoi cell tessellation. This is easily produced with the Point Volume Interpolator filter. This filter takes points from a dataset, treats them like a point cloud, and interpolates them on a regular grid defined in space. One of the interpolating kernels is a Voronoi Kernel, so you can use the resulting grid to query values and approximate the Voronoi tessellation.
This is good. But somehow this doesn’t work very well for my data, which has at least 400,000 points and flashes warnings. SPH on the other hand gives values which make sense.
That’s true. I’m not sure why you are getting warnings, but the volume interpolator is only as good as the resolution you set the grid to. With 400,000 points you would probably need to crank the resolution up to an unreasonable size to capture the shape of the cells with any accuracy.
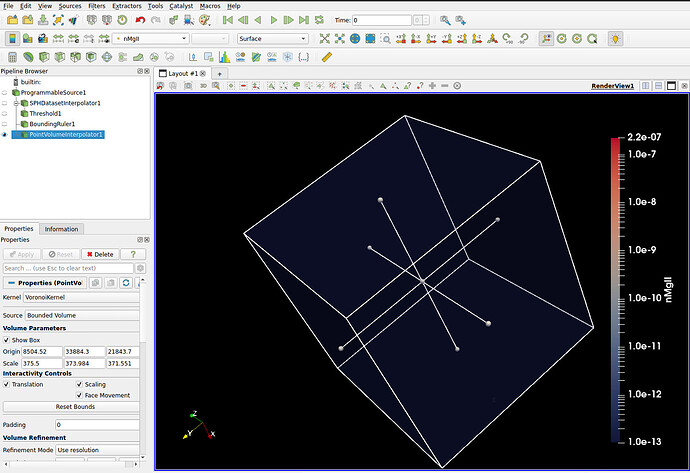
Here are my data files and the ParaView state. Please let me know if you can reproduce the warnings and perhaps fix this. I am using a programmable source to read the data, where you need to change the location to point to the data file I am attaching after loading the state.
halo-prop-nMgII_ID=110-clipped.npy|attachment (1.7 MB)
paraview11.pvsm (581.5 KB)
I can’t seem to replicate your warnings (in ParaView 3.12.0-RC1). Once I update your state to point to my directory, I get no warnings.
The Voronoi filter with Point Volume interpolator works fine for you?
Seems to work OK. I got some warnings when I tried to do volume rendering due to the way the rendering properties were set up, but adding the Point Volume Interpolator to the output of your Programmable Source seemed to work just fine.
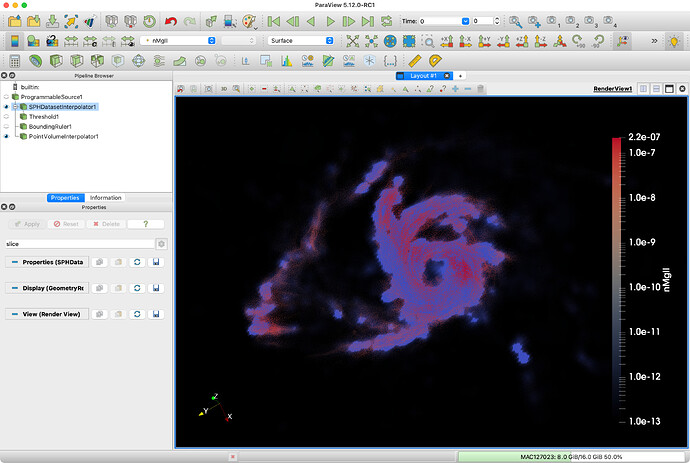
Point Volume Interpolator with volume rendering is exactly what was giving me the warning as well. Also, the slice plots after the Point Volume Interpolator give results with values and features that don’t make much sense and, intuitively, seem incorrect by just looking at the point data.
Like I said, I don’t think the warning has anything to do with the Point Volume Interpolator. It’s a problem with the precision of the rendering, as discussed here: cannot use log scale due to opengl texture problem
About the results not making sense, I don’t know enough about your work to know about that. I still suspect it is a precision problem. The two data sets look reasonably close given the coarseness of the grid.
point-volume-interp.pvsm (791.5 KB)
@Kenneth_Moreland Thanks a lot! I was making a surface representation which was causing this.