I have a workflow using Dask-MPI to move data distributed across multiple nodes into VTK for rendering. The source data is a regular grid fp32 supernova dataset that is pre-partitioned by Dask. Each MPI rank gets one partition of the data, stored in numpy format, which it uses to locally build a vtkImageData. I then use the vtkImageData object to initialize a trivial producer. I’m getting hung up on setting the data extents properly, however.
Here’s a snippet of the python code I’m using for processing the data, with just two ranks for testing. ar is the numpy array storing the volume data (originally 3D), dims are the dimensions of the local chunk, and wdims are the dimensions of the entire volume. For this simple case, I’m just splitting along the z-axis. Persistent objects like producers and paraview.simple are stored in the wrapping class for this code.
print(“Rank”, self.rank, “has array with local/global dims”, dims, wdims)
ar = np.reshape(ar, dims[0]*dims[1]*dims[2])
ext = [0,dims[0]-1, 0,dims[1]-1, max(self.rank*dims[2]-1,0),(self.rank+1)*dims[2]-1]
wext = [0,wdims[0]-1, 0,wdims[1]-1, 0,wdims[2]-1]
vtkimg = vtk.vtkImageData()
vtkimg.Initialize()
vtkimg.SetExtent(ext)
vtkimg.SetSpacing([1,1,1])
#set the extent for the whole dataset
vi = vtk.vtkInformation()
vtkimg.CopyInformationToPipeline(vi)
vi.Set(vtk.vtkStreamingDemandDrivenPipeline.WHOLE_EXTENT(), wext[0],wext[1],wext[2],wext[3],wext[4],wext[5])
vtkimg.CopyInformationFromPipeline(vi)
varnm = 'E' #'E' is entropy for this data
vtkarr = vtknp.numpy_to_vtk(ar)
vtkarr.SetName(varnm)
vtkimg.GetPointData().AddArray(vtkarr)
vtkimg.GetPointData().SetScalars(vtkarr)
self.TP = self.pvs.TrivialProducer()
self.TP.GetClientSideObject().SetOutput(vtkimg)
self.TP.UpdatePipeline()
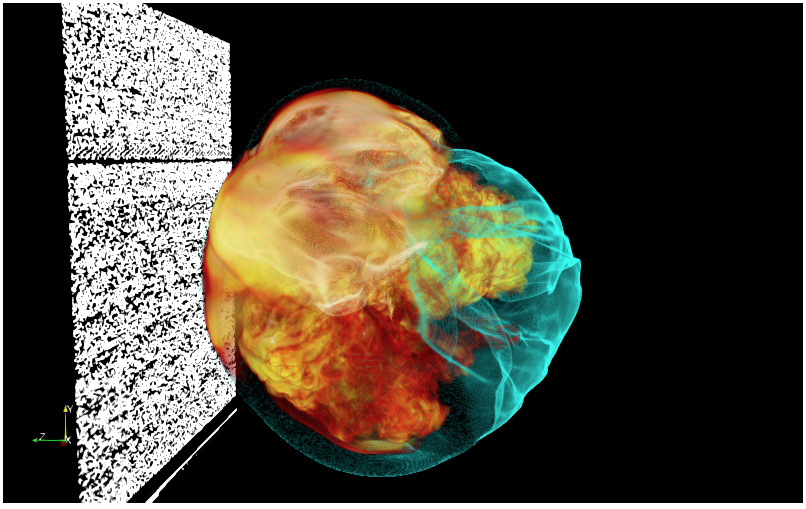
This mostly works, but I’m getting junk data aligned with the split axis (see the image below) or sometimes a segfault. That looks like a pretty clear off-by-one case to me. I’ve verified that the local and global dimensions are correct–(432,432,216) and (432,432,432), respectively–so I must be specifying them incorrectly in VTK. I notice that I’m not getting a visible seam between partitions, however, which I would expect if data at the partition boundary were wrong or missing ghost cells. Unfortunately, search results for this topic are a bit of a minefield and bring up a lot of hits for old, obsolete methods.