Hello ParaView community,
first off, I am fairly new in the ParaView cosmos but am highly excited seeing the capabilities it offers especially with regards to my CFD simulation data that I have acquired and am trying to post-process.
I am trying to run ParaView5.6 on a TACC cluster, primarily in serial (though I have run it with 2 MPI processes as well) as the visualization node I’m using offers plenty (2.1TB to be exact) memory, but am experiencing issues when loading large datasets with the XDMF2 loader. Specifically, I’m talking about an HDF5 file containing XYZ + 5 conservative fluid variables on ~ 5 billion grid points which equates to a 309GB solution file for one time instance.
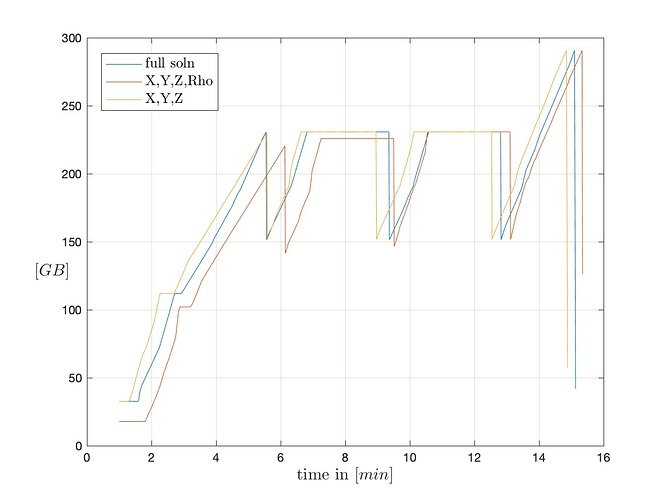
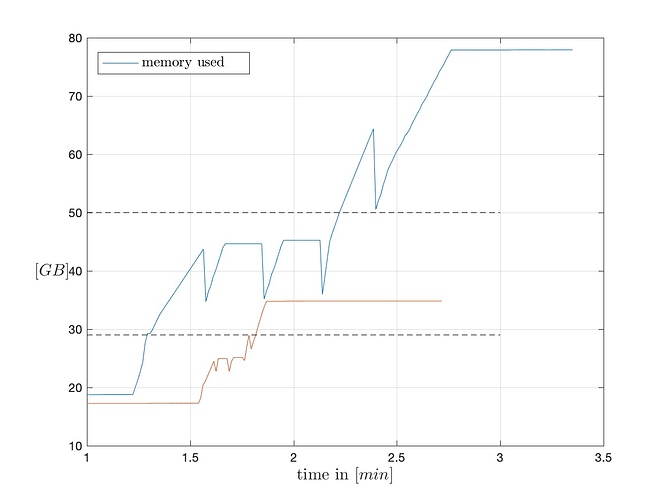
Problem being, that when I try to load the dataset with ParaView run in GUI mode, it tries to load the data into memory for a few minutes before it crashes without throwing an error in the command line (it’s run through Software Rasterizer (swr) ). When running a “free -m” during this process, I can see that ParaView is a) trying to load the data and then crashing and b) crashing before fully loading the data (see attached plot). Also, I tried reading in only the grid + 1 flow variable (density) as well as only the grid by specifying the XDMF file accordingly, but the crash happens almost identically.
I have tried swapping out the solution file with a lower resolution case that is 1/8th of the size and it works just fine (see plot below). This seemingly is a problem that arises solely because of the solution file size, but cannot be related to a lack of sufficient memory as I have plenty available before it crashes.
There are obviously different ways to go about this, e.g. dividing solution files into multiple slabs and loading subsequently, or using multi-grids options. Subsampling the solutions files (every other point) has been helpful in visualizing but that means a) I throw away picture quality and b) requires an extra computationally expensive and storage intensive step to the procedure.
Has anyone experienced similar issues with XFDM2 files associate with large datasets and/or has an idea what could be happening and possibly how to avoid this?
Thanks in advance for your help,
Fabian
Comparison loading full solution vs. XYZ+density vs. XYZ only:
Comparison loading full solution for subsampled solution files (every second / third grid point):