Hello everyone,
I have a 3D parallel unstructured code. If I run the code in parallel with lets say 32 processors, I get 32 vtu files with corresponding grid(local+ghost) data and field data. I also have a pvtu file to visualize those. My first problem was, when I load the pvtu file into paraview, I get a distorted plot with all the partitions overlapping(please see before.png). Later, when i use Clean to grid and Clean cells to grid filter, I get a proper grid without the overlaps(see after.png). But when i use these filters my field data is removed.
I sincerely request anyone in this forum to help me with this problem. Also, kindly let me know if there are any other better ways of plotting partitioned files. I am using a 3D wing case for this problem.
Regards,
Srikanth
Short answer: use Clean to Grid only. Do not use Clean Cells to Grid.
When you render an unstructured grid with the default surface representation, ParaView does so by extracting the external surface. The automatic surface extraction used is quick and lazy and ignores ghost cells. Normally, this has no visual consequence because you are usually looking at the geometry from the outside. However, if I understand your images correctly, your mesh is actually of the volume around a wing and you have to zoom inside the geometry to see the wing itself. In this case, you see the ghost surfaces that the representation has not bothered to remove.
Running Clean to Grid should solve the problem by merging duplicate points on the local compute node. It should also pass the field data correctly. The Clean Cells to Grid filter is what is removing your field data. it probably shouldn’t (and I raised a bug about it), but you shouldn’t need this filter anyway. Clean Cells to Grid is about removing degenerate cells, and you probably don’t have any.
Hi Kenneth,
Thanks for you response!
Yes, you are right. The wing is inside the geometry. If I only use clean to grid filter, the repeating ghost cells are removed.
As you can see, the field data is still intact. How do you think I should proceed from here.
Forgive my ignorance on this, I have been running the parallel code for sometime now. I am finding it hard to process the large output and dealing with ghost cells and what not.
Do you think there is a better way to deal with this problem?
Regards,
Srikanth
“I’m finding it hard to process” is a pretty open question, so it might be better to ask a more specific question.
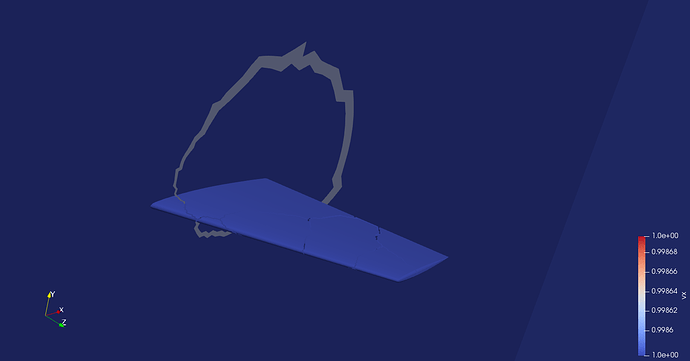
However, looking closely at the image you posted, I notice that there appear to be cracks in the wing. I’m going to make an educated guess (since I don’t have direct access to your data) and suppose that (1) this is at least one of the issues you are referring to and (2) that the cause of these cracks are the ghost cells.
What I’m pretty sure is happening is that where there are ghost cells you have an overlap of cells: one of the “real” cell and at least one of the duplicated “ghost” cell. Because the ghost cells share points (after the grid is “cleaned”) they share all their faces and therefore none of their faces are classified as “external”.
What I think is happening is that ParaView is not properly identifying what is a ghost cell and what is not. Your data set should have a cell array named vtkGhostType of type UInt8 that specifies which cells are ghost cells. Does your data set have that?
I just added vtkGhostType(setting 0 for local and 1 for ghost cells) to the dataset and now as soon as i load the .pvtu file, i get a dataset which clears the ghost information( without performing clean to grid filter. Please see below,
Yes, I reckon the cause of the cracks is due to the ghost cells too. Also, I am uploading my .vtu and .pvtu files to this thread. Please let me know if you think there are any problems with the format of the vtu files.
data.tar.xz (2.5 MB)
I want to thank you for your valuable insights.
Regards,
Srikanth