Dear ParaView experts,
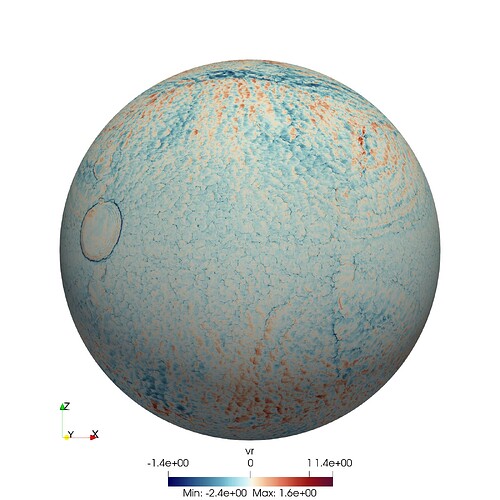
Context: I am working on a very large astrophysical simulation. Our parallel code splits the computational domain into hundreds of thousands of small, independent, structured grids (typical size 24^3 gridpoints). In one variant of that simulation setup, these meshes are slightly tilted with respect to each other (see first figure). This is to cover the surface of the star.
Pipeline: To visualise such a setup, I first convert all tiny meshes from native format to individual *.vts files using the PyVista StructuredGrid() & save() functions. Next, I manually write the content of the *.vtm file from the list of all the *.vts files. The content of this file looks something like this:
<?xml version="1.0"?>
<VTKFile type="vtkMultiBlockDataSet" version="1.0" byte_order="LittleEndian" header_type="UInt32" compressor="vtkZLibDataCompressor">
<vtkMultiBlockDataSet>
<Block index="1">
<DataSet index="10513" time="84.400000" name="Block-10513" file="VB96f_0211/VB96f_0211_10513.vts"/>
<DataSet index="10519" time="84.400000" name="Block-10519" file="VB96f_0211/VB96f_0211_10519.vts"/>
....
<DataSet index="10543" time="84.400000" name="Block-10543" file="VB96f_0211/VB96f_0211_10543.vts"/>
</Block>
</vtkMultiBlockDataSet>
</VTKFile>
I have no problem reading this file in ParaView; despite this setup being made out of small cubes, a spherical cut makes a nice, smooth preview of the surface (see second figure), so
Kudos to ParaView!! ![]()
Issues and questions: The only problem this pipeline has is performance. By default I am firing up ./mpiexec -np 8 -map-by numa ./pvserver --disable-xdisplay-test --force-offscreen-rendering and I see that data is nicely distributed, but initial I/O takes forever (~40 min for our small case i.e., 10k of small meshes, until I see initial outline frame), and during readout, it looks like only one core is working.
For comparison, I have also exported small grids (still using PyVista) to multiple *.vtk files and I fussed them into one big *.vtk with this script:
import glob
from vtk import *
reader = vtkStructuredGridReader()
append = vtkAppendFilter()
filenames = glob.glob('VB6*.vtk')
# filenames = filenames[0:2]
for file in filenames:
reader.SetFileName(file)
reader.Update()
structured = vtkStructuredGrid()
structured.ShallowCopy(reader.GetOutput())
append.AddInputData(structured)
append.Update()
# writer = vtkPolyDataWriter()
writer = vtkUnstructuredGridWriter()
writer.SetFileName('output.vtk')
writer.SetInputData(append.GetOutput())
writer.Write()
This seems to read in a bit faster (even if on one core!). Also slicing, it is faster in that case. This leads me to believe that maybe I am using the wrong format and there is some obvious flaw in my pipeline.
Questions: Is there a better format for what I aim to do? I was trying to fuse only some groups of my small meshes into N *.vtk files; next, I was trying to build an XML/*.vts file with their list, but ParaView didn’t like it. I have also tried to add more blocks i.e,. <Block index="n"> to my *.vts but that made I/O even slower.
Any advice and guidance is welcome since, with every iteration, we are adding more and more small elements, and visualisation is more and more troublesome with the pipeline we have right now.