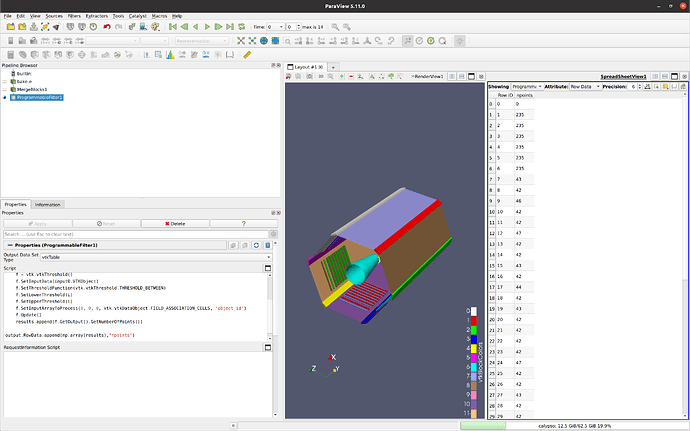
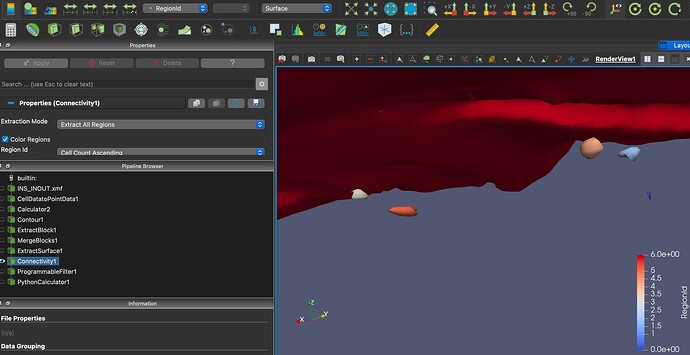
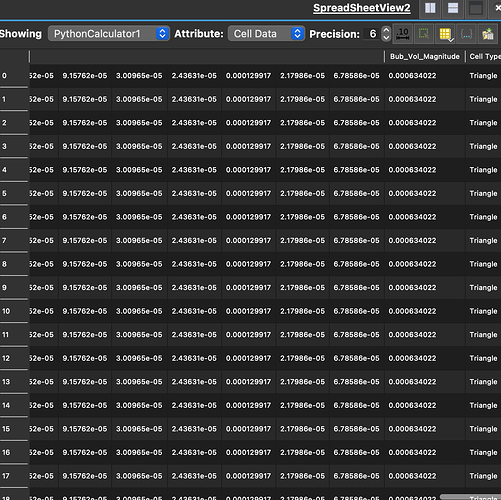
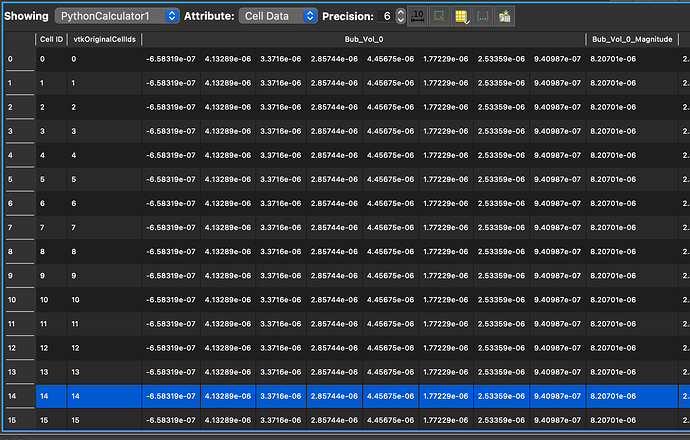
Thanks, almost there I think 
I tweaked the code based on your suggestion.
import vtk
import numpy as np
k=5 # number of regions total
input0 = inputs[0]
for i in range(k):
f = vtk.vtkThreshold()
f.SetInputData(input0.VTKObject)
f.SetThresholdFunction(vtk.vtkThreshold.THRESHOLD_BETWEEN)
f.SetLowerThreshold(i)
f.SetUpperThreshold(i)
f.SetInputArrayToProcess(0, 0, 0, vtk.vtkDataObject.FIELD_ASSOCIATION_CELLS, 'RegionId')
f.Update()
input1 = f.GetOutput()
numTriangles = input1.GetNumberOfCells()
volumeArray = np.empty([numTriangles,k], dtype=np.float64)
for j in range(numTriangles):
cell = input1.GetCell(j)
p1 = input1.GetPoint(cell.GetPointId(0))
p2 = input1.GetPoint(cell.GetPointId(1))
p3 = input1.GetPoint(cell.GetPointId(2))
p4 = [1.058105,2.47113,-0.209546] # use barycenter of surface for better results.
#p4 = [1.0,1.0,-1.0]
volumeArray[j,i] = vtk.vtkTetra.ComputeVolume(p1,p2,p4,p3)
output.CellData.append(volumeArray[:,i], "Bub_Vol")
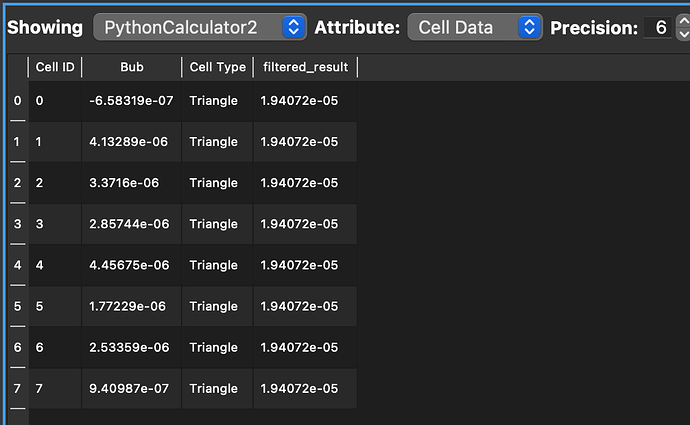
It appears I am getting something when I look at spreadsheet view (see below), but it doesnt make sense. the rows are all the same. In the end I just want an array of the total volumes (5 values in my case), I dont really care about the individual contributions, just need the sum of the array if this makes sense.
this is the error.
Traceback (most recent call last):
File "<string>", line 22, in <module>
File "<string>", line 17, in RequestData
File "/Applications/ParaView-5.10.1.app/Contents/Python/vtkmodules/numpy_interface/dataset_adapter.py", line 130, in __getattr__
return getattr(self.VTKObject, name)
AttributeError: 'vtkmodules.vtkCommonDataModel.vtkUnstructuredGrid' object has no attribute 'RowData'
Traceback (most recent call last):
File "/Applications/ParaView-5.10.1.app/Contents/Python/paraview/detail/calculator.py", line 203, in execute
retVal = compute(inputs, expression, ns=variables)
File "/Applications/ParaView-5.10.1.app/Contents/Python/paraview/detail/calculator.py", line 146, in compute
retVal = eval(subEx, globals(), mylocals)
File "<string>", line 1, in <module>
NameError: name 'Bub_Vol' is not defined
Traceback (most recent call last):
File "<string>", line 22, in <module>
File "<string>", line 17, in RequestData
File "/Applications/ParaView-5.10.1.app/Contents/Python/vtkmodules/numpy_interface/dataset_adapter.py", line 130, in __getattr__
return getattr(self.VTKObject, name)
AttributeError: 'vtkmodules.vtkCommonDataModel.vtkUnstructuredGrid' object has no attribute 'RowData'
Traceback (most recent call last):
File "/Applications/ParaView-5.10.1.app/Contents/Python/paraview/detail/calculator.py", line 203, in execute
retVal = compute(inputs, expression, ns=variables)
File "/Applications/ParaView-5.10.1.app/Contents/Python/paraview/detail/calculator.py", line 146, in compute
retVal = eval(subEx, globals(), mylocals)
File "<string>", line 1, in <module>
NameError: name 'Bub_Vol' is not defined
Traceback (most recent call last):
File "<string>", line 22, in <module>
File "<string>", line 17, in RequestData
File "/Applications/ParaView-5.10.1.app/Contents/Python/vtkmodules/numpy_interface/dataset_adapter.py", line 130, in __getattr__
return getattr(self.VTKObject, name)
AttributeError: 'vtkmodules.vtkCommonDataModel.vtkUnstructuredGrid' object has no attribute 'RowData'
Traceback (most recent call last):
File "<string>", line 22, in <module>
File "<string>", line 17, in RequestData
File "/Applications/ParaView-5.10.1.app/Contents/Python/vtkmodules/numpy_interface/dataset_adapter.py", line 130, in __getattr__
return getattr(self.VTKObject, name)
AttributeError: 'vtkmodules.vtkCommonDataModel.vtkUnstructuredGrid' object has no attribute 'RowData'
Traceback (most recent call last):
File "<string>", line 22, in <module>
File "<string>", line 17, in RequestData
File "/Applications/ParaView-5.10.1.app/Contents/Python/vtkmodules/numpy_interface/dataset_adapter.py", line 130, in __getattr__
return getattr(self.VTKObject, name)
AttributeError: 'vtkmodules.vtkCommonDataModel.vtkUnstructuredGrid' object has no attribute 'RowData'
Traceback (most recent call last):
File "<string>", line 22, in <module>
File "<string>", line 18, in RequestData
AttributeError: 'vtkmodules.vtkFiltersCore.vtkThreshold' object has no attribute 'GetNumberOfCells'
Traceback (most recent call last):
File "<string>", line 22, in <module>
File "<string>", line 18, in RequestData
AttributeError: 'vtkmodules.vtkFiltersCore.vtkThreshold' object has no attribute 'GetNumberOfCells'
Traceback (most recent call last):
File "<string>", line 22, in <module>
File "<string>", line 18, in RequestData
AttributeError: 'vtkmodules.vtkFiltersCore.vtkThreshold' object has no attribute 'GetNumberOfCells'
Traceback (most recent call last):
File "<string>", line 22, in <module>
File "<string>", line 18, in RequestData
AttributeError: 'vtkmodules.vtkFiltersCore.vtkThreshold' object has no attribute 'GetNumberOfCells'
Traceback (most recent call last):
File "/Applications/ParaView-5.10.1.app/Contents/Python/paraview/detail/calculator.py", line 203, in execute
retVal = compute(inputs, expression, ns=variables)
File "/Applications/ParaView-5.10.1.app/Contents/Python/paraview/detail/calculator.py", line 146, in compute
retVal = eval(subEx, globals(), mylocals)
File "<string>", line 1
sum(
^
SyntaxError: unexpected EOF while parsing
Traceback (most recent call last):
File "<string>", line 22, in <module>
File "<string>", line 18, in RequestData
AttributeError: 'vtkmodules.vtkFiltersCore.vtkThreshold' object has no attribute 'GetNumberOfCells'
Traceback (most recent call last):
File "/Applications/ParaView-5.10.1.app/Contents/Python/paraview/detail/calculator.py", line 203, in execute
retVal = compute(inputs, expression, ns=variables)
File "/Applications/ParaView-5.10.1.app/Contents/Python/paraview/detail/calculator.py", line 146, in compute
retVal = eval(subEx, globals(), mylocals)
File "<string>", line 1
sum(
^
SyntaxError: unexpected EOF while parsing
Traceback (most recent call last):
File "/Applications/ParaView-5.10.1.app/Contents/Python/paraview/detail/calculator.py", line 203, in execute
retVal = compute(inputs, expression, ns=variables)
File "/Applications/ParaView-5.10.1.app/Contents/Python/paraview/detail/calculator.py", line 146, in compute
retVal = eval(subEx, globals(), mylocals)
File "<string>", line 1, in <module>
NameError: name 'Bub_Vol' is not defined
Traceback (most recent call last):
File "<string>", line 22, in <module>
File "<string>", line 18, in RequestData
AttributeError: 'vtkmodules.vtkFiltersCore.vtkThreshold' object has no attribute 'GetNumberOfCells'
Traceback (most recent call last):
File "/Applications/ParaView-5.10.1.app/Contents/Python/paraview/detail/calculator.py", line 203, in execute
retVal = compute(inputs, expression, ns=variables)
File "/Applications/ParaView-5.10.1.app/Contents/Python/paraview/detail/calculator.py", line 146, in compute
retVal = eval(subEx, globals(), mylocals)
File "<string>", line 1, in <module>
NameError: name 'Bub_Vol' is not defined
ERROR: In /opt/glr/paraview/paraview-ci/build/superbuild/paraview/src/VTK/Common/ExecutionModel/vtkDemandDrivenPipeline.cxx, line 760
vtkPVDataRepresentationPipeline (0x600005de00f0): Input for connection index 0 on input port index 0 for algorithm vtkGeometryRepresentationWithFaces(0x16fd06fa0) is of type vtkTable, but a vtkDataSet is required.
ERROR: In /opt/glr/paraview/paraview-ci/build/superbuild/paraview/src/VTK/Common/ExecutionModel/vtkDemandDrivenPipeline.cxx, line 760
vtkPVDataRepresentationPipeline (0x600005d0ab20): Input for connection index 0 on input port index 0 for algorithm vtkGeometryRepresentation(0x12fe67250) is of type vtkTable, but a vtkDataSet is required.
ERROR: In /opt/glr/paraview/paraview-ci/build/superbuild/paraview/src/VTK/Common/ExecutionModel/vtkDemandDrivenPipeline.cxx, line 760
vtkPVDataRepresentationPipeline (0x600005d05680): Input for connection index 0 on input port index 0 for algorithm vtkPVGridAxes3DRepresentation(0x164245290) is of type vtkTable, but a vtkCompositeDataSet is required.
Traceback (most recent call last):
File "<string>", line 22, in <module>
File "<string>", line 18, in RequestData
AttributeError: 'vtkmodules.vtkFiltersCore.vtkThreshold' object has no attribute 'GetNumberOfCells'
Traceback (most recent call last):
File "/Applications/ParaView-5.10.1.app/Contents/Python/paraview/detail/calculator.py", line 203, in execute
retVal = compute(inputs, expression, ns=variables)
File "/Applications/ParaView-5.10.1.app/Contents/Python/paraview/detail/calculator.py", line 146, in compute
retVal = eval(subEx, globals(), mylocals)
File "<string>", line 1, in <module>
NameError: name 'Bub_Vol' is not defined
Traceback (most recent call last):
File "/Applications/ParaView-5.10.1.app/Contents/Python/paraview/detail/calculator.py", line 203, in execute
retVal = compute(inputs, expression, ns=variables)
File "/Applications/ParaView-5.10.1.app/Contents/Python/paraview/detail/calculator.py", line 146, in compute
retVal = eval(subEx, globals(), mylocals)
File "<string>", line 1, in <module>
NameError: name 'Bub_Vol' is not defined
0.004158140694556545
Traceback (most recent call last):
File "/Applications/ParaView-5.10.1.app/Contents/Python/paraview/detail/calculator.py", line 203, in execute
retVal = compute(inputs, expression, ns=variables)
File "/Applications/ParaView-5.10.1.app/Contents/Python/paraview/detail/calculator.py", line 146, in compute
retVal = eval(subEx, globals(), mylocals)
File "<string>", line 1, in <module>
NameError: name 'Bub_Vol' is not defined
nan
Traceback (most recent call last):
File "/Applications/ParaView-5.10.1.app/Contents/Python/paraview/detail/calculator.py", line 203, in execute
retVal = compute(inputs, expression, ns=variables)
File "/Applications/ParaView-5.10.1.app/Contents/Python/paraview/detail/calculator.py", line 146, in compute
retVal = eval(subEx, globals(), mylocals)
File "<string>", line 1, in <module>
NameError: name 'Bub_Vol' is not defined
1.71592883706e-312
Traceback (most recent call last):
File "/Applications/ParaView-5.10.1.app/Contents/Python/paraview/detail/calculator.py", line 203, in execute
retVal = compute(inputs, expression, ns=variables)
File "/Applications/ParaView-5.10.1.app/Contents/Python/paraview/detail/calculator.py", line 146, in compute
retVal = eval(subEx, globals(), mylocals)
File "<string>", line 1, in <module>
NameError: name 'Bub_Vol' is not defined
Traceback (most recent call last):
File "/Applications/ParaView-5.10.1.app/Contents/Python/paraview/detail/calculator.py", line 203, in execute
retVal = compute(inputs, expression, ns=variables)
File "/Applications/ParaView-5.10.1.app/Contents/Python/paraview/detail/calculator.py", line 146, in compute
retVal = eval(subEx, globals(), mylocals)
File "<string>", line 1, in <module>
NameError: name 'Bub_Vol' is not defined
Traceback (most recent call last):
File "/Applications/ParaView-5.10.1.app/Contents/Python/paraview/detail/calculator.py", line 203, in execute
retVal = compute(inputs, expression, ns=variables)
File "/Applications/ParaView-5.10.1.app/Contents/Python/paraview/detail/calculator.py", line 146, in compute
retVal = eval(subEx, globals(), mylocals)
File "<string>", line 1, in <module>
NameError: name 'Bub_Vol' is not defined
Traceback (most recent call last):
File "/Applications/ParaView-5.10.1.app/Contents/Python/paraview/detail/calculator.py", line 203, in execute
retVal = compute(inputs, expression, ns=variables)
File "/Applications/ParaView-5.10.1.app/Contents/Python/paraview/detail/calculator.py", line 146, in compute
retVal = eval(subEx, globals(), mylocals)
File "<string>", line 1, in <module>
NameError: name 'Bub_Vol' is not defined```