I’ve had some more time to experiment with different ways of writing the output format, and with some help from @mwestphal, I think I have figured out what I was doing wrong back in December and how to fix it.
My initial problem: When writing partitioned datasets, each partition is processed individually. For some filters to work correctly, information about cells from neighboring partitions is needed. One such filter is the Contour filter. If only the data from one partition is processed, the resulting isosurface might not be continuous across partitions. This is because the filter averages cell data to the points of the mesh to interpolate the surface. To get the same value at points shared between partitions, all surrounding cell data values are needed. This information is provided through ghost cells, which I did not include in my output file:
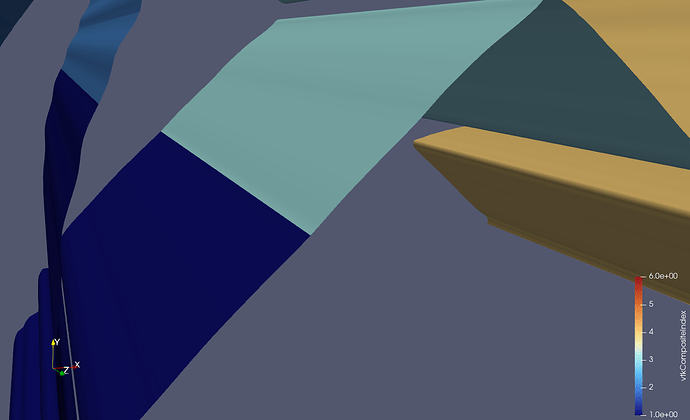
This picture shows a different (smaller) dataset than the one that I originally posted. The domain is partitioned into two partitions, of which the first one is shown. I added some gaps between the cells to make it more clear what I am talking about. The dark blue cells are the inner cells, and the orange and light blue cells are boundary cells. The inner cells are regular 3D cells, while the boundary cells are 2D cells used to specify boundary conditions (not relevant here). If you wanted to compute isosurfaces for a dataset without ghost cells, the only way to get good results is to use the Ghost Cells filter in ParaView (important detail: turn on Generate Global Ids in the settings). This will generate the missing ghost cells for you, at the cost of additional computation during post-processing. Originally, this filter did not work for me, probably because I was using a ParaView binary from my package manager, but after switching to an official release binary, everything worked as expected. The additional cost comes from ParaView having to send and receive the ghost cell data to and from the individual partitions (MPI ranks). I don’t have any performance measurements on this, but I would suspect that this is actually not too bad, compared to the effort of doing this yourself…
Doing this yourself: In my case, the data comes from a cell-based finite volume code, in which I already have to perform communication between the MPI ranks. For the solution procedure, I need to compute the flux across each face of the mesh. A face is the triangle or quadrangle between two cells. This means that for cells on the boundary of a partition, I need to have access to the cell values on the neighboring partition. For this exchange to work, each partition keeps track of a set of halo cells, which, after the exchange, hold the cell value of the neighboring partition (the red cells):
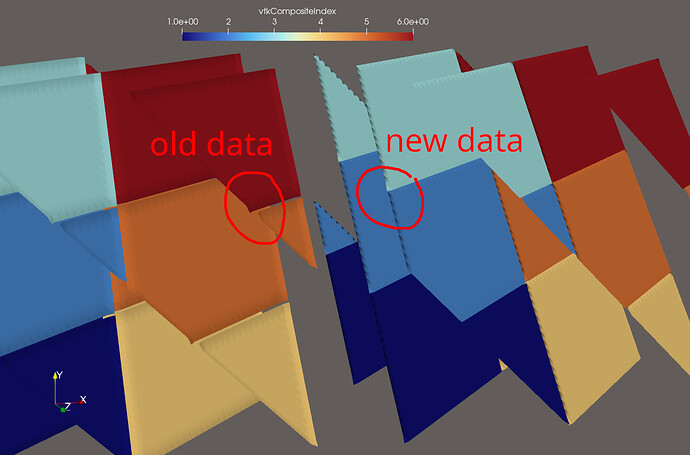
After adding these cells to the output file and marking them as DUPLICATECELL (=1) in a special data array called “vtkGhostType”, the situation improved, and it looked like this would resolve my issue. However, the dataset I was using for my analysis was quasi-structured, meaning the cells that are adjacent to all the points of one partition are the same cells that are adjacent to the cells of that partition (except for the edges and corners). Therefore, it seemed like it was working, prompting me to mark this issue as solved, while this was only a special case and not a general solution.
A general solution: As I mentioned at the very top (long ago, I know…), the Contour filter needs to have access to all the cells that are adjacent to all the points within one partition. Only then will the average value (most likely weighted average…) be the same at every shared point. This is what such an output file looks like:
As you can see, there are considerably more halo cells in the output, and crucially, every cell which is made up of at least one node from that partition. This is the form that outputs must have in order for filters like Contour to work as expected. With this dataset, no further processing is needed in ParaView. In fact, if you are not explicitly looking for these ghost cells, you would never know that they are there.
The procedure for determining adjacent cells will depend on your code/setup. For me, I was already using ParMETIS to compute the dual (cell-to-cell) of my cell graph (cell-to-node). The ParMETIS_V3_PartKway function takes as an input argument ncommonnodes, which is set to 3 to obtain the dual that you need for a finite volume method (in 2D it would be set to 2). With a little bit of pre- and post-processing, you can set this argument to 1, to obtain a graph that tells you for each cell which other cells share at least one node with it. This is the information that I used to compute the send/receive information for the complete set of halo cells. Of course, you only need to exchange the information of the complete set, when you are writing outputs. For regular finite volume operations, it is enough to only exchange the cells obtained from ncommonnodes = 3. If you reorder the halo cells such that these cells come first, this works as if these extra cells were not even there.
Alternative solutions: Writing halo cells yourself is a bit of a hassle and really only necessary if you are dealing with large datasets. What you can do instead is write the file in parallel, but remap the local node indices to global ones. For this to work, you only need to keep the unique global index of every mesh point around. For me, this information comes directly from the mesh generator, so it should not be difficult to obtain. When doing this, the output file will look to ParaView like it was written from a single rank, which means you can completely omit any halo cells. I think there is some limit to the number of points/cells that ParaView can handle per single partition. I did not specifically go looking for it, but I’ve had it crash when my cell counts approached INT32_MAX, so this method will only work for meshes with less than about 2e9 points/cells.
Here are the datasets from the plots:
-
No ghost cells:
no_ghost_cells.hdf (145.0 KB)
-
Face-connected ghost cells:
face_connected.hdf (150.2 KB)
-
Point-connected ghost cells:
point_connected.hdf (164.5 KB)
-
Single partition:
single_partition.hdf (189.7 KB)
(If you are asking yourself why the single partition dataset is the largest, the answer is that I am using int64_t for integers instead of int32_t.)