Hello,
I have cell data which correspond to grains. I would like to outline the edges that share 2 different grain IDs, a.k.a. grain boundaries.
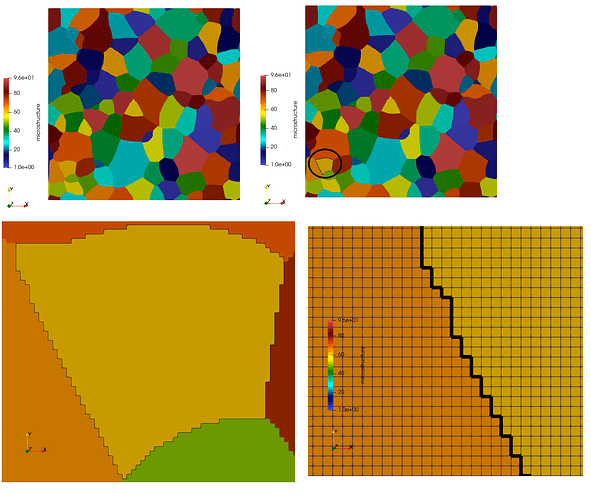
I did an example (see picture) with 1 grain. I did:
threshold of 1 grain + feature edge view representation
I am not willing to do this by brute force (going 1 by 1). I am wondering if there is a simple solution to do this.
Many thanks,
mwestphal
August 4, 2020, 2:45pm
2
This is very nice looking ! Thanks for sharing.
As for your question, I’m afraid there is no better way to do that currently.
An alternative could be to use the “DigitalRockPhysics” plugin and its “ExplodeMaterialClusters” filters.https://gitlab.kitware.com/paraview/paraview/-/merge_requests/2184
SC_REN
January 28, 2021, 2:53pm
3
Hi, I had the same problem. I hope you find a solution.
Finally, I used python shell to loop on the thereshold-FeatureEdge procedure to extract the grain boundaries.
BR,
Hello,
what we did was:
calculate the points where the boundaries are.
Construct ‘quad’ elements in VTK using the previous points.
Visualize them as feature edges
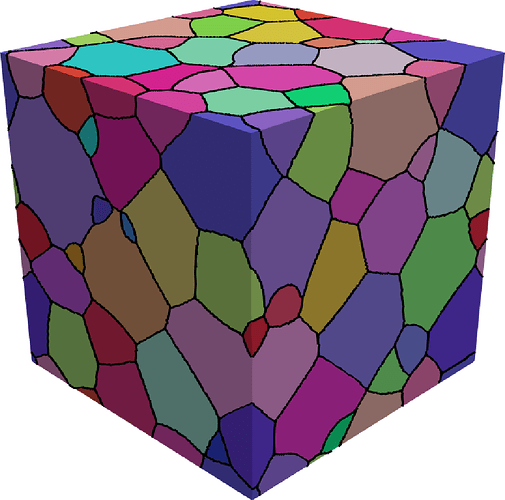
We also upgrade it to 3D. Also vectorize it, avoiding loops and making it a bit more efficient.
If needed, let me know and i can send the code.
Cheers,
F.Niessen
May 6, 2021, 2:26pm
5
Dear Fran,
I would be interested in the code, I am trying to visualize a similar structure. That would be very appreciated.
Best wishes
getGrainBoundaries_DAMASK.py (1.8 KB)
Hello, here the code. It uses functions from modules of DAMASK ( Files · v3.0.0-alpha3 · damask / DAMASK · GitLab (mpie.de) ).
Something like this would be the work flow.
Cheers,
N.S
June 20, 2022, 5:07am
8
Thanks for the very valuable code!
By the way, can Grid.load() read a vtr file, or if it’s a vti, I’m having trouble getting an error.
Currently, Grid.load reads both, vti and vtr.
N.S
June 20, 2022, 6:50am
10
Thank you for your prompt reply.
I am having trouble with the following error.
I thought Grid.load() was GriddataFomats, is that correct?