Hi all,
based on the previous post (Stresses from ANSYS .rst are displayed as NaN - #2 by Kenneth_Moreland) I have noticed the following problem.
With pymapdl_reader only the individual components of the stress tensor are extracted. And that in the nodes. ParaView links these stress components under Magnitude, but the program apparently uses the L2 norm:
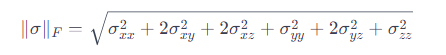
What I actually need for my analysis is the von Mises stress instead L2 norm:
![]()
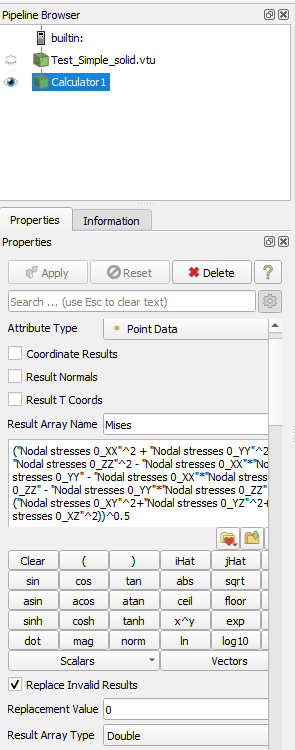
I have tried to generate this equivalent stress with a calculator:
("Nodal stresses 0_XX"^2 + "Nodal stresses 0_YY"^2 + "Nodal stresses 0_ZZ"^2 -
"Nodal stresses 0_XX"*"Nodal stresses 0_YY" - "Nodal stresses 0_XX"*"Nodal stresses 0_ZZ" - "Nodal stresses 0_YY"*"Nodal stresses 0_ZZ" +
3 * ("Nodal stresses 0_XY"^2+"Nodal stresses 0_YZ"^2+"Nodal stresses 0_XZ"^2))^0.5
However, I get a completely incorrect distribution of stress in the body:
It should be mentioned at this point that the numerical values in the scale are correct. So the calculator has probably calculated the stresses correctly, but it is likely to be the visualization that is failing.
I am using ParaView 5.12.0-RC2. A .vtu file with the stresses and nodes can be found in this post:
https://discourse.paraview.org/t/stresses-from-ansys-rst-are-displayed-as-nan/13836/2
I need please help with the visualization of the stresses with a Calculator.
Many thanks in advance
ingenium