@Andy_Bauer @utkarsh.ayachit @mwestphal @Francois_Mazen @nicolas.vuaille or anyone experienced in using catalyst, please help me out.
I wanted to use Catalyst for in-situ visualization in a C astrophysical hydrodynamics code called PLUTO. I have been able to successfully use catalyst by modifying the CFullExample code provided with minimal changes in the original source code of PLUTO.
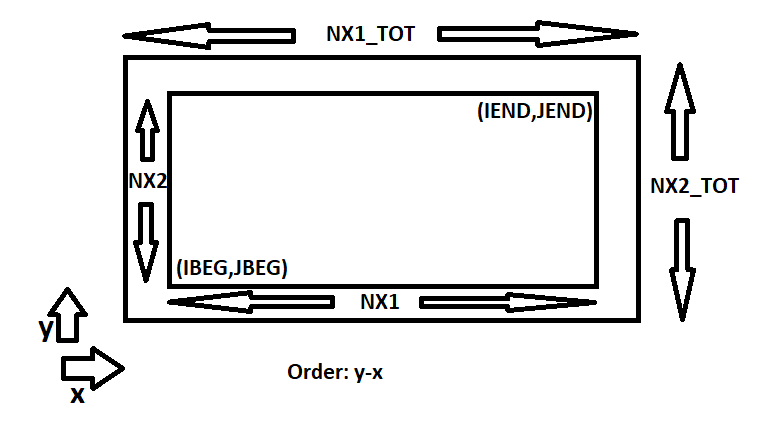
The CFullExample simulation code uses a grid and field data that is structured in such a way that is compatible with Catalyst (vtk). Unfortunately, PLUTO simulation grid and field data arrangement is not natively compatible with Catalyst. Therefore, I had to copy the grid data and the field data to the compatible structures as given in the Example code. These structures are:
typedef struct Grid
{
uint64_t NumberOfPoints;
uint64_t NumberOfCells;
double* Points;
int64_t* Cells;
} Grid;
typedef struct Attributes
{
// A structure for generating and storing point and cell fields.
// Velocity is stored at the points and pressure is stored
// for the cells.
double* Velocity;
float* Pressure;
Grid* GridPtr;
} Attributes;
Therefore, each time there is a update of grid or field data, I had to copy the data from PLUTO simulation and update the members of these data structures. Then I’m able to get the correct results when using the code provided in CatalystAdaptor.h.
For example, the following line in CatalystAdaptor.h assumes that the (point) data, which is already structured in vtk compatible format, is present at the memory location of the pointer to the array grid->Points. As a result, I had to create a vtk compatible grid->Points array which is properly structured by copying data from native PLUTO arrays.
Copying every time from PLUTO arrays before Catalyst accesses the data makes the code slow and also creates redundant memory footprint containing the same data.
conduit_node_set_path_external_float64_ptr_detailed(mesh, "coordsets/coords/values/x",
/*data=*/grid->Points, /*num_elements=*/grid->NumberOfPoints, /*offset=*/0,
/*stride=*/3 * sizeof(double), /*element_bytes=*/sizeof(double),
/*endianness=*/CONDUIT_ENDIANNESS_DEFAULT_ID);
The way out I think would be to make every element of grid->Points array (similarly other grid/field data) to not contain values but pointers to memory addresses of the required values from the original native PLUTO arrays. This eliminates the need for copying during every update (as the simulation already updates them). However, this requires rather than direct reading of data at the location of grid->Points, dereferencing the addresses contained by every element in grid->Points.
This is where I’m stuck because using the functions in CatalystAdaptor.h doesn’t seem to do this job.
Please help. I’m new to Catalyst.
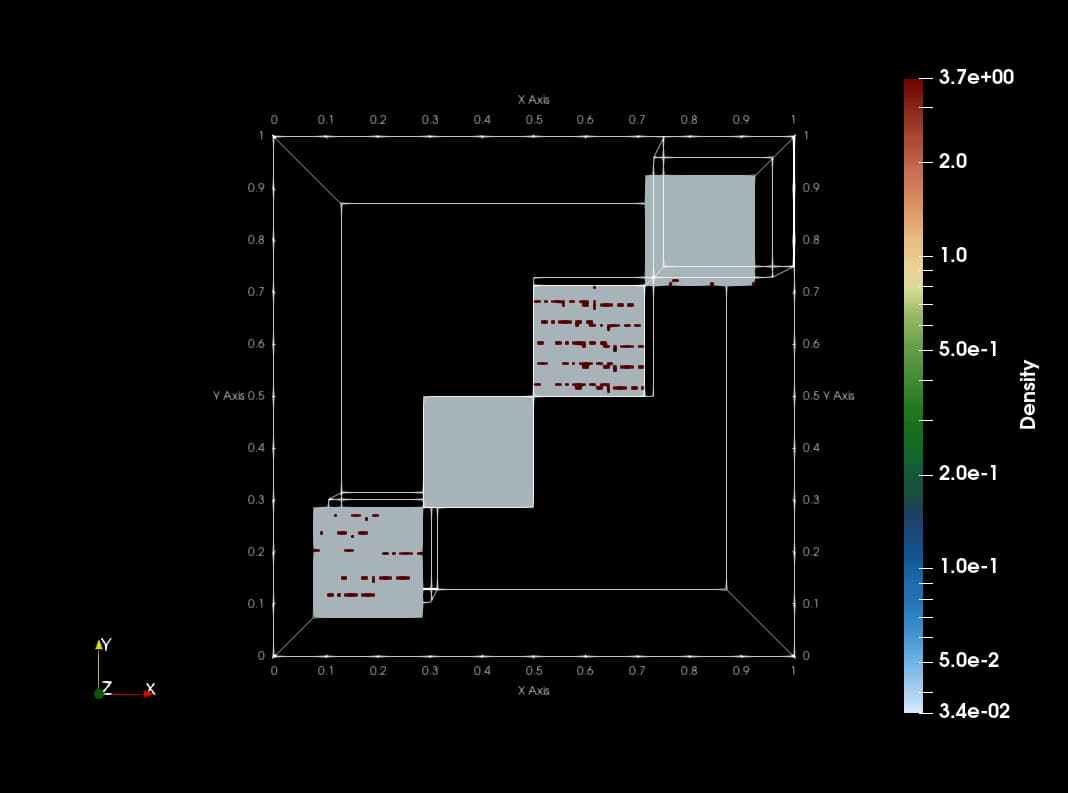
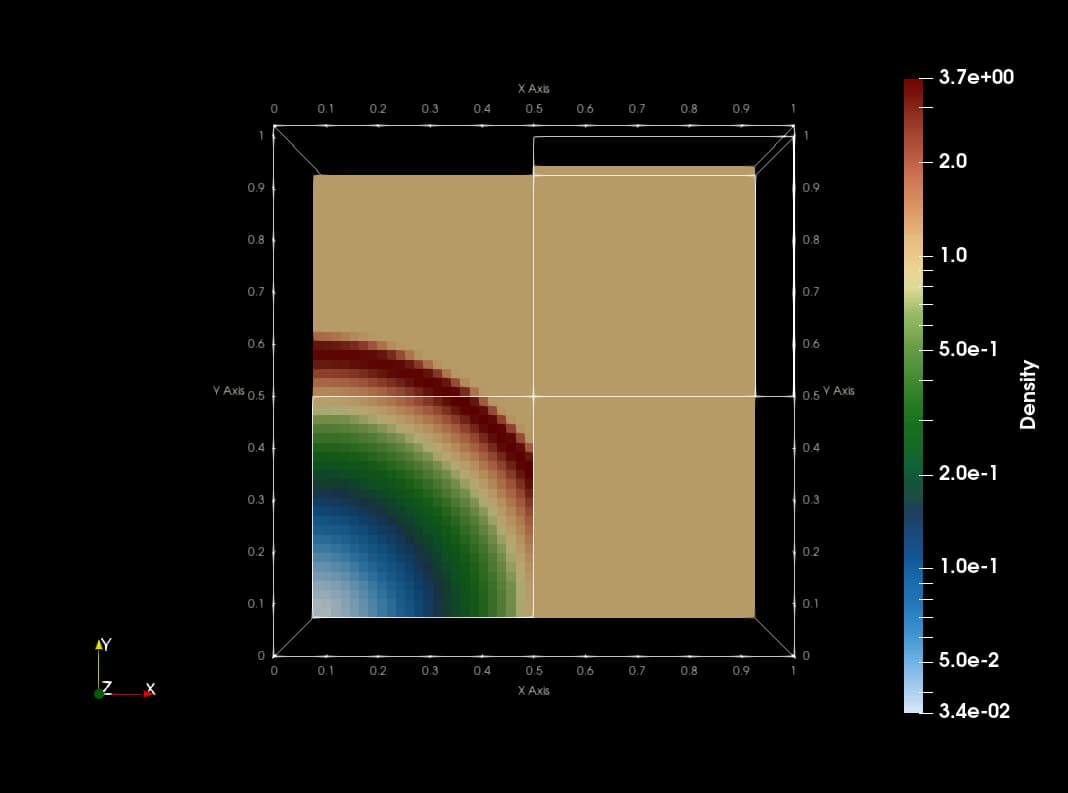
This animation is made with png dumps from a run of the PLUTO code. The code uses Catalyst to create slices of blastwave formation in real time. The current implementation copies data from native PLUTO arrays to vtk compatible array structures which makes the code slow with redundant memory footprint.